Share/CLEF eHealth 2013 TASK 3 Results - QUTTOPSIG
This page summarises the results obtained from your submissions. Here, you can find the mean performance of your submissions, for all standard trec_eval measures and for nDCG at different ranks. For this first year, the Share/CLEF eHealth 2013 TASK 3 built result pools from your submissions considering the top 10 documents ranked by your baseline system (run 1), and the highest priority run that used the discharge summaries (run 2) and the highest priority run that did not used the discharge summaries (run 5). As a consequence:
- the primary measure for this year is precision at 10 (P@10),
- the secondary measure is Normalised Discounted Cumulative Gain at rank 10 (ndcg_cut_10).
Evaluation with standard trec_eval metrics
These results have been obtained with the binary relevance assessment, i.e. qrels.clef2013ehealth.1-50-test.bin.final.txt, and trec_eval 9.0 as distributed by NIST. Trec_eval was ran as follows:
./trec_eval -c -M1000 qrels.clef2013ehealth.1-50-test.bin.final.txt runName
QUTTOPSIG.1.3.noadd
runid all Topsig num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 1478 map all 0.1969 gm_map all 0.0536 Rprec all 0.2344 bpref all 0.3053 recip_rank all 0.5019 iprec_at_recall_0.00 all 0.5495 iprec_at_recall_0.10 all 0.4506 iprec_at_recall_0.20 all 0.3462 iprec_at_recall_0.30 all 0.2518 iprec_at_recall_0.40 all 0.2159 iprec_at_recall_0.50 all 0.1774 iprec_at_recall_0.60 all 0.1401 iprec_at_recall_0.70 all 0.1105 iprec_at_recall_0.80 all 0.0812 iprec_at_recall_0.90 all 0.0461 iprec_at_recall_1.00 all 0.0052 P_5 all 0.3520 P_10 all 0.3540 P_15 all 0.3053 P_20 all 0.2760 P_30 all 0.2327 P_100 all 0.1330 P_200 all 0.0844 P_500 all 0.0499 P_1000 all 0.0296
QUTTOPSIG.2.3.noadd
runid all Topsig num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 1478 map all 0.1965 gm_map all 0.0532 Rprec all 0.2344 bpref all 0.3058 recip_rank all 0.4569 iprec_at_recall_0.00 all 0.5327 iprec_at_recall_0.10 all 0.4683 iprec_at_recall_0.20 all 0.3472 iprec_at_recall_0.30 all 0.2518 iprec_at_recall_0.40 all 0.2159 iprec_at_recall_0.50 all 0.1774 iprec_at_recall_0.60 all 0.1401 iprec_at_recall_0.70 all 0.1105 iprec_at_recall_0.80 all 0.0812 iprec_at_recall_0.90 all 0.0461 iprec_at_recall_1.00 all 0.0052 P_5 all 0.3520 P_10 all 0.3560 P_15 all 0.3053 P_20 all 0.2760 P_30 all 0.2327 P_100 all 0.1330 P_200 all 0.0844 P_500 all 0.0499 P_1000 all 0.0296
QUTTOPSIG.3.3.noadd
runid all Topsig num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 1443 map all 0.1841 gm_map all 0.0430 Rprec all 0.2197 bpref all 0.2987 recip_rank all 0.3870 iprec_at_recall_0.00 all 0.4738 iprec_at_recall_0.10 all 0.4361 iprec_at_recall_0.20 all 0.3354 iprec_at_recall_0.30 all 0.2727 iprec_at_recall_0.40 all 0.2193 iprec_at_recall_0.50 all 0.1603 iprec_at_recall_0.60 all 0.1300 iprec_at_recall_0.70 all 0.0956 iprec_at_recall_0.80 all 0.0745 iprec_at_recall_0.90 all 0.0385 iprec_at_recall_1.00 all 0.0059 P_5 all 0.3040 P_10 all 0.3240 P_15 all 0.3000 P_20 all 0.2600 P_30 all 0.2173 P_100 all 0.1258 P_200 all 0.0815 P_500 all 0.0476 P_1000 all 0.0289
QUTTOPSIG.4.3.noadd
runid all TopISSL num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 445 map all 0.0340 gm_map all 0.0008 Rprec all 0.0619 bpref all 0.1562 recip_rank all 0.1350 iprec_at_recall_0.00 all 0.1518 iprec_at_recall_0.10 all 0.0948 iprec_at_recall_0.20 all 0.0657 iprec_at_recall_0.30 all 0.0442 iprec_at_recall_0.40 all 0.0335 iprec_at_recall_0.50 all 0.0298 iprec_at_recall_0.60 all 0.0180 iprec_at_recall_0.70 all 0.0129 iprec_at_recall_0.80 all 0.0108 iprec_at_recall_0.90 all 0.0000 iprec_at_recall_1.00 all 0.0000 P_5 all 0.0720 P_10 all 0.0560 P_15 all 0.0480 P_20 all 0.0510 P_30 all 0.0580 P_100 all 0.0300 P_200 all 0.0202 P_500 all 0.0155 P_1000 all 0.0089
QUTTOPSIG.5.3.noadd
runid all Topsig num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 1443 map all 0.1821 gm_map all 0.0427 Rprec all 0.2197 bpref all 0.2984 recip_rank all 0.3853 iprec_at_recall_0.00 all 0.4685 iprec_at_recall_0.10 all 0.4227 iprec_at_recall_0.20 all 0.3387 iprec_at_recall_0.30 all 0.2760 iprec_at_recall_0.40 all 0.2193 iprec_at_recall_0.50 all 0.1603 iprec_at_recall_0.60 all 0.1300 iprec_at_recall_0.70 all 0.0956 iprec_at_recall_0.80 all 0.0745 iprec_at_recall_0.90 all 0.0385 iprec_at_recall_1.00 all 0.0059 P_5 all 0.3040 P_10 all 0.3240 P_15 all 0.3000 P_20 all 0.2600 P_30 all 0.2173 P_100 all 0.1258 P_200 all 0.0815 P_500 all 0.0476 P_1000 all 0.0289
QUTTOPSIG.6.3.noadd
runid all Topsig num_q all 50 num_ret all 50000 num_rel all 1858 num_rel_ret all 1186 map all 0.0724 gm_map all 0.0055 Rprec all 0.0875 bpref all 0.2533 recip_rank all 0.1761 iprec_at_recall_0.00 all 0.2186 iprec_at_recall_0.10 all 0.1496 iprec_at_recall_0.20 all 0.1269 iprec_at_recall_0.30 all 0.1110 iprec_at_recall_0.40 all 0.0817 iprec_at_recall_0.50 all 0.0697 iprec_at_recall_0.60 all 0.0539 iprec_at_recall_0.70 all 0.0446 iprec_at_recall_0.80 all 0.0377 iprec_at_recall_0.90 all 0.0257 iprec_at_recall_1.00 all 0.0019 P_5 all 0.0880 P_10 all 0.0860 P_15 all 0.0973 P_20 all 0.0950 P_30 all 0.0840 P_100 all 0.0682 P_200 all 0.0541 P_500 all 0.0375 P_1000 all 0.0237
Evaluation with nDCG
These results have been obtained with the graded relevance assessment, i.e. qrels.clef2013ehealth.1-50-test.graded.final.txt, and trec_eval 9.0 as distributed by NIST.To obtain nDCG at different ranks, trec_eval was ran as follows:
./trec_eval -c -M1000 -m ndcg_cut qrels.clef2013ehealth.1-50-test.graded.final.txt runName
QUTTOPSIG.1.3.noadd
ndcg_cut_5 all 0.3185 ndcg_cut_10 all 0.3293 ndcg_cut_15 all 0.3082 ndcg_cut_20 all 0.2975 ndcg_cut_30 all 0.2875 ndcg_cut_100 all 0.3521 ndcg_cut_200 all 0.3816 ndcg_cut_500 all 0.4155 ndcg_cut_1000 all 0.4346
QUTTOPSIG.2.3.noadd
ndcg_cut_5 all 0.3090 ndcg_cut_10 all 0.3242 ndcg_cut_15 all 0.3038 ndcg_cut_20 all 0.2942 ndcg_cut_30 all 0.2852 ndcg_cut_100 all 0.3504 ndcg_cut_200 all 0.3798 ndcg_cut_500 all 0.4136 ndcg_cut_1000 all 0.4328
QUTTOPSIG.3.3.noadd
ndcg_cut_5 all 0.2651 ndcg_cut_10 all 0.2846 ndcg_cut_15 all 0.2829 ndcg_cut_20 all 0.2695 ndcg_cut_30 all 0.2635 ndcg_cut_100 all 0.3296 ndcg_cut_200 all 0.3554 ndcg_cut_500 all 0.3838 ndcg_cut_1000 all 0.4076
QUTTOPSIG.4.3.noadd
ndcg_cut_5 all 0.0672 ndcg_cut_10 all 0.0619 ndcg_cut_15 all 0.0586 ndcg_cut_20 all 0.0622 ndcg_cut_30 all 0.0726 ndcg_cut_100 all 0.0904 ndcg_cut_200 all 0.1019 ndcg_cut_500 all 0.1187 ndcg_cut_1000 all 0.1298
QUTTOPSIG.5.3.noadd
ndcg_cut_5 all 0.2617 ndcg_cut_10 all 0.2820 ndcg_cut_15 all 0.2805 ndcg_cut_20 all 0.2671 ndcg_cut_30 all 0.2609 ndcg_cut_100 all 0.3265 ndcg_cut_200 all 0.3523 ndcg_cut_500 all 0.3808 ndcg_cut_1000 all 0.4045
QUTTOPSIG.6.3.noadd
ndcg_cut_5 all 0.0814 ndcg_cut_10 all 0.0778 ndcg_cut_15 all 0.0852 ndcg_cut_20 all 0.0877 ndcg_cut_30 all 0.0896 ndcg_cut_100 all 0.1362 ndcg_cut_200 all 0.1753 ndcg_cut_500 all 0.2065 ndcg_cut_1000 all 0.2328
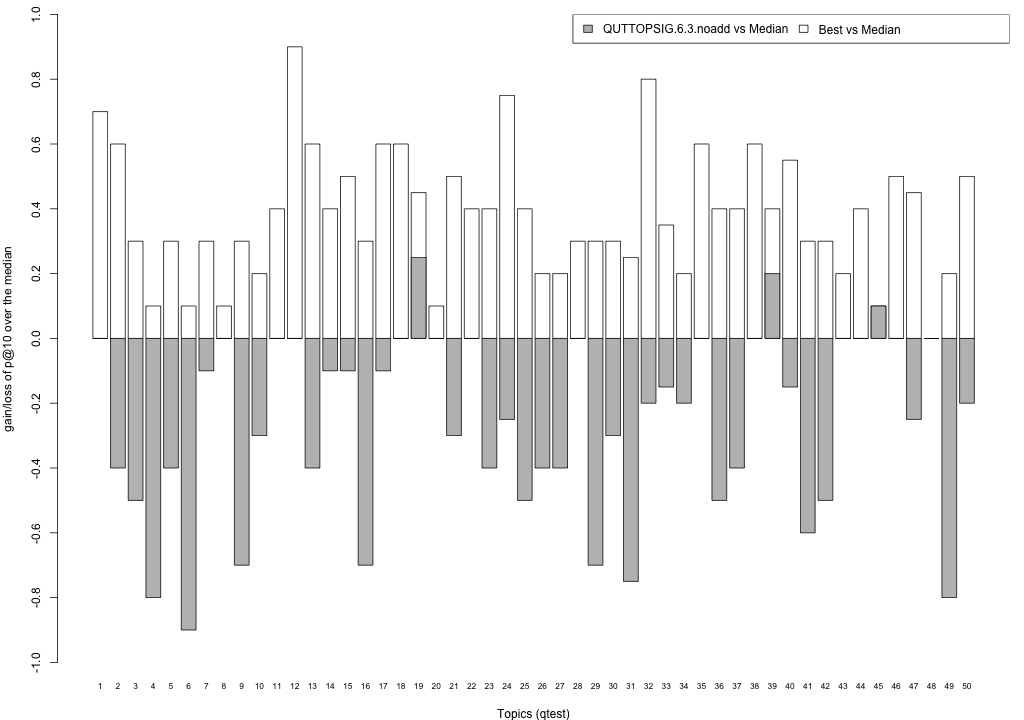
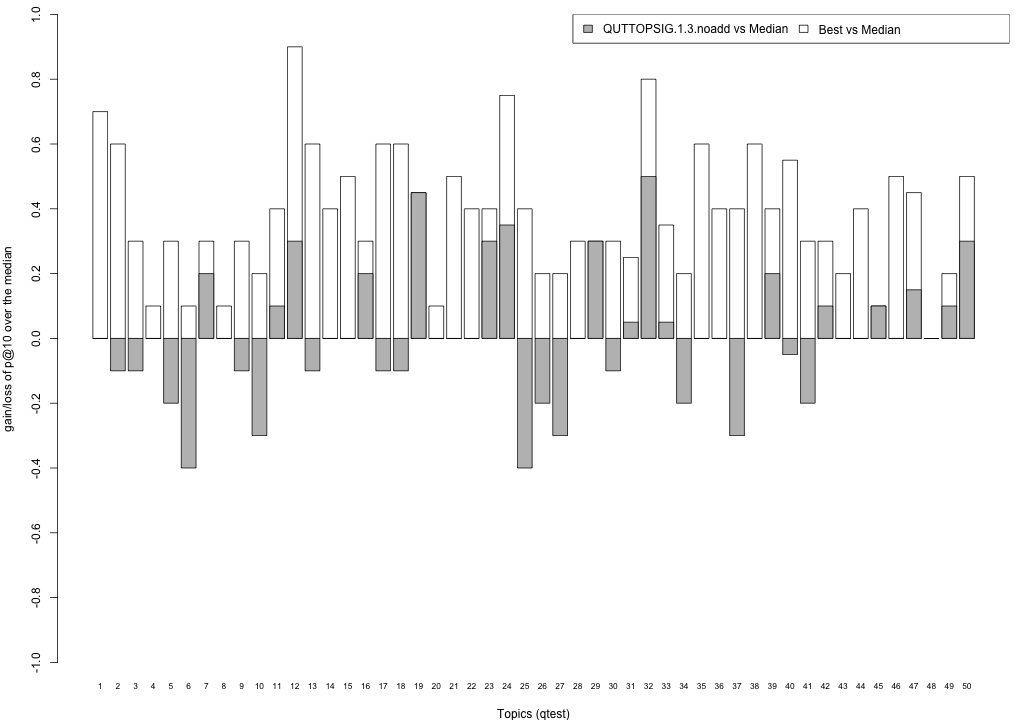
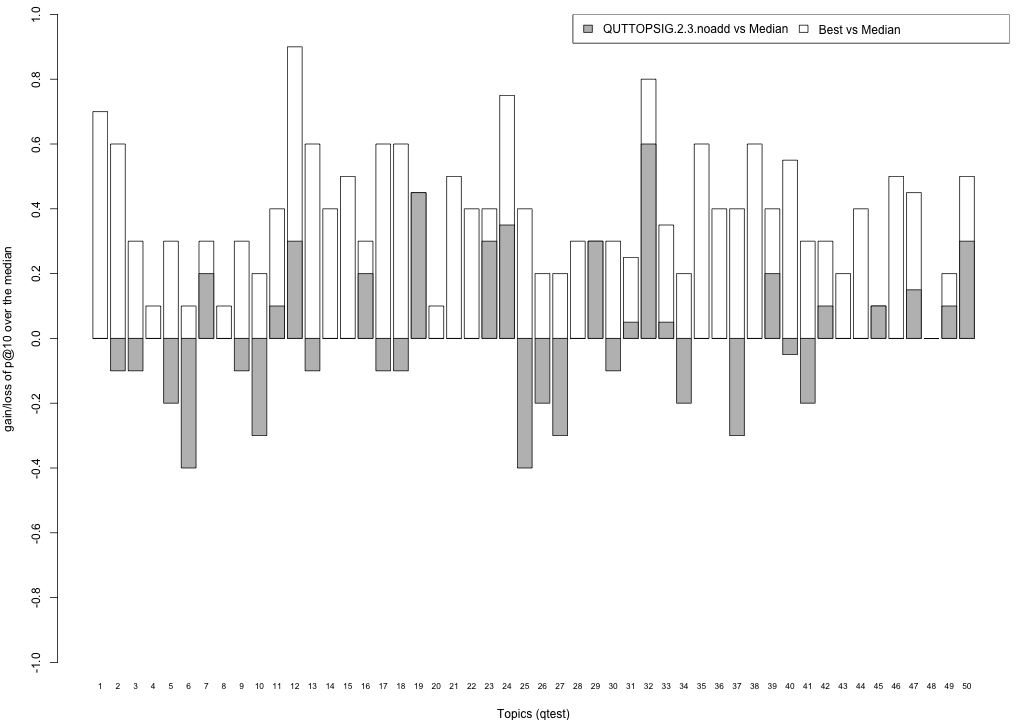
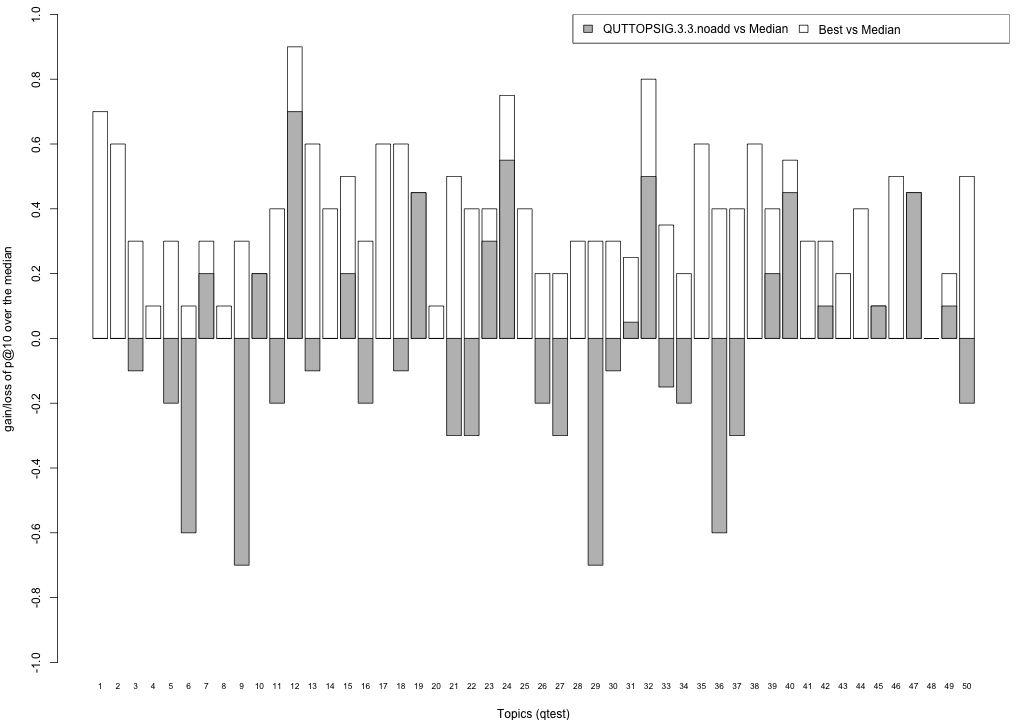
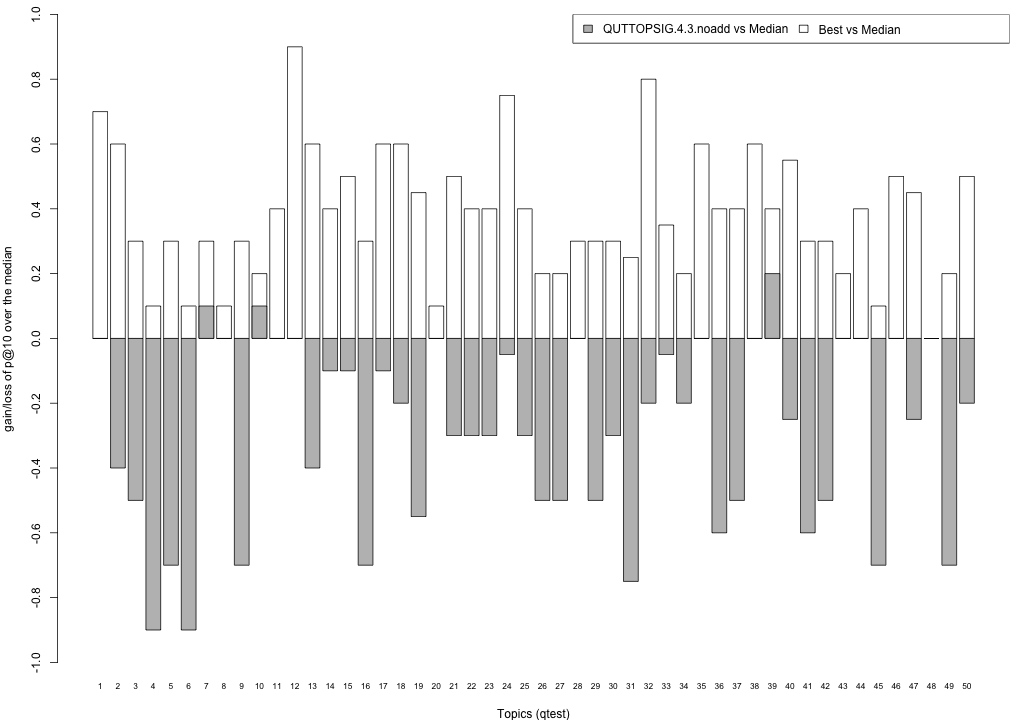
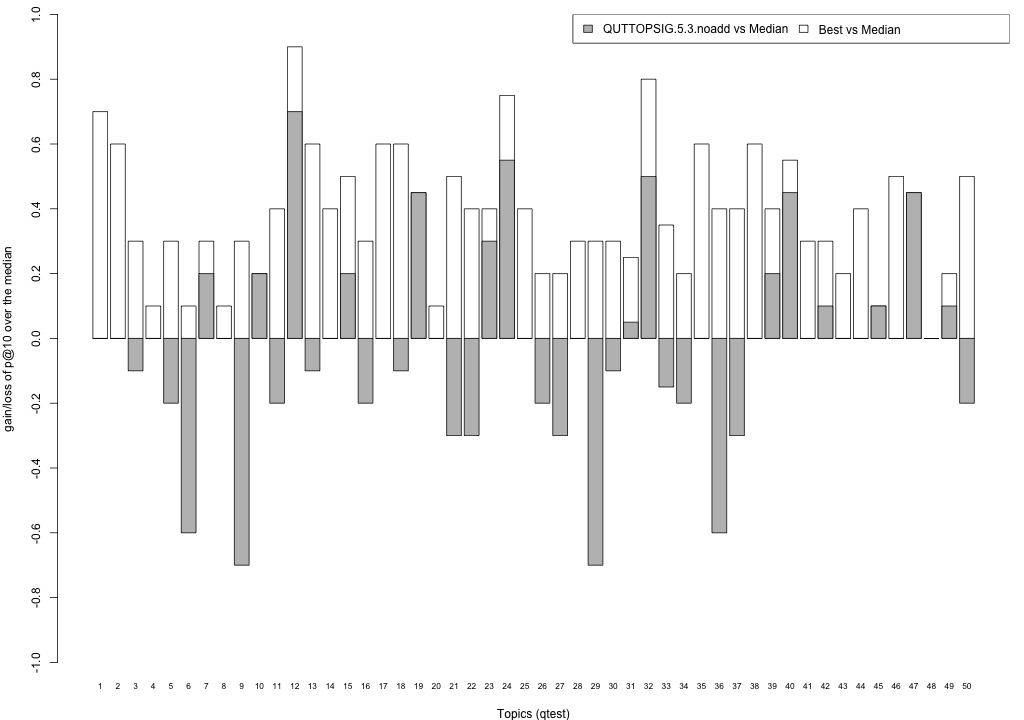
Plots P@10
The plots below compare each of your runs against the median and best performance (p@10) across all systems submitted to CLEF for each query topic. In particular, for each query, the height of a bar represents the gain/loss of your system and the best system (for that query) over the median system. The height of a bar in then given by:
grey bars: height(q) = your_p@10(q) - median_p@10(q) white bars: height(q) = best_p@10(q) - median_p@10(q)
QUTTOPSIG.1.3.noadd
QUTTOPSIG.2.3.noadd
QUTTOPSIG.3.3.noadd
QUTTOPSIG.4.3.noadd
QUTTOPSIG.5.3.noadd
QUTTOPSIG.6.3.noadd